CGPACK > NOV-2016
7-NOV-2016: A report on TAU and CrayPAT
paper.pdf - profiling of CGPACK and CGPACK+ParaFEM multi-scale models with TAU and CrayPAT.
8-NOV-2016: Checking new NetCDF writer
Luis has written unit test
testACF.f90
which writes CA data in NetCDF format.
In addition this test also does timing.
The user prepares several directories with
different lsf settings.
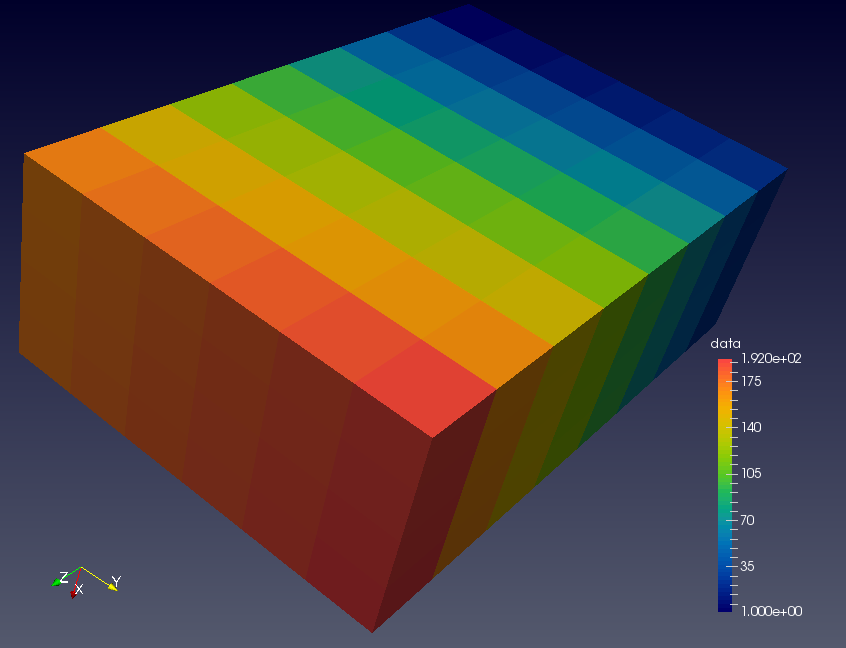
Below is the output file written in NetCDF
read into ParaView.
The cells are initialised with the image number.
The code was run on 192 images, so the colour bar shows
cubes with colours from dark blue (image 1) to red (image 192).
The file was saved with *.ncdf extension.
In was read as ParaView "netCDF files generic and CF conventions".

Since this seems to work, cut release 2.7.
ncdump
is a useful utility.
It can convert a binary NetCDF file into
plain text.
This is particularly useful for extracting metadata.
$ ncdump -cs netcdf.dat
netcdf netcdf {
dimensions:
x = 464 ;
y = 930 ;
z = 1392 ;
variables:
int data(z, y, x) ;
data:_Storage = "contiguous" ;
data:_Endianness = "little" ;
data:_NoFill = "true" ;
// global attributes:
:_Format = "netCDF-4" ;
data:
}
9-NOV-2016: NetCDF timings
testACF.f90
has been updated to automatically create dirs
with different lfs stripe counts and sizes.
The following values have been studied:
character( len=maxlen), dimension(9) :: stripe_count = (/ &
"-c-1 ", "-c0 ", "-c1 ", "-c4 ", "-c8 ", &
"-c16 ", "-c20 ", "-c32 ", "-c40 " /)
character( len=maxlen), dimension(7) :: stripe_size = (/ &
"-S1m ", "-S2m ", "-S4m ", "-S8m ", "-S16m", &
"-S32m", "-S64m" /)
Files are saved in directores with names formed by stripe count and size. Each file has about 10^9 cells, i.e. about 4.5GB.
Data from 8 Archer nodes, sorted by IO rate in descending order. Pure MPI/IO peaks at 4GB/s. NetCDF gives only just above 1GB/s.
13-NOV-2016: Luis added an HDF5 writer
Luis added an HDF5 writer in module
cgca_m2hdf5.f90.
I updated test
testACF.f90
to dump the model also in HDF5.
HDF5 file cannot be read directly into ParaView,
because the metadata contained in HDF5 file does
not fully describe it.
I had to write a simple xdmf metadata file for it:
hdf5.xdmf.
The xdmf file is given to ParaView.
The necessary metadata can be extracted from the hdf5 file
with
h5ls
and
h5dump
h5dump
utilities:
$ h5ls hdf5.dat
IntArray Dataset {1392, 930, 930}
$ h5dump -H hdf5.dat
HDF5 "hdf5.dat" {
GROUP "/" {
DATASET "IntArray" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 1392, 930, 930 ) / ( 1392, 930, 930 ) }
}
}
}
Below is the CA brick where cell values are image numbers. This was run on 8 ARcher cores (480 images).
